Plant trichomes vary in shape, size, and structure and are key to plant defense against biotic and abiotic stresses such as insect herbivory, high temperature, and UV radiation. While Arabidopsis serves as a model for unicellular trichome studies, tomatoes possess seven types of multicellular trichomes with both glandular and nonglandular features. Earlier studies identified Woolly (Wo), HD8, and Hair (H/H2) genes as critical for tomato trichome development, yet the upstream regulators and cooperative molecular interactions remained elusive. Due to these gaps, researchers needed to investigate how zinc finger proteins coordinate with other transcription factors to orchestrate multicellular trichome formation in tomato.
Researchers from Seoul National University, Chonnam National University, and Gyeongsang National University have uncovered a new regulatory pathway controlling tomato trichome development. The study(DOI: 10.1093/hr/uhaf008), published on April 1, 2025, in Horticulture Research, reports that SlH3 and SlH4 act as redundant C2H2 zinc finger proteins promoting multicellular trichome formation and elongation by interacting with Wo. Through gene-editing, microscopy, and transcriptomic analyses, the team demonstrated that these proteins form homo- and heterodimers and boost Wo-mediated activation of downstream genes such as Wox3b and MX1.
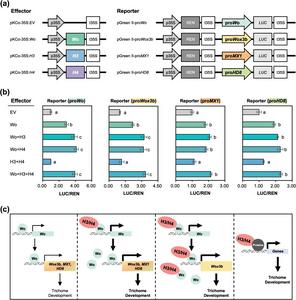
Using CRISPR-Cas9 technology, the team generated single and double knockout lines of SlH3 and SlH4. While single mutants showed no significant change, the double mutants exhibited markedly fewer glandular trichomes (Types I, VI, VII) and more nonglandular trichomes (Types III, V). Additionally, the Type I trichomes were shorter due to reduced stalk cell numbers, revealing that SlH3 and SlH4 redundantly promote trichome formation and elongation. Protein assays confirmed that SlH3 and SlH4 form both homo- and heterodimers and physically interact with Wo, suggesting the formation of a cooperative transcriptional complex. RNA sequencing revealed downregulation of Wo, Wox3b, MX1, and HD8 in double mutants, genes all known to contribute to trichome differentiation. Dual-luciferase assays further demonstrated that SlH3/SlH4–Wo complexes strongly enhanced transcriptional activity from Wo and Wox3b promoters but not from MX1 or HD8, indicating gene-specific modulation. These findings establish SlH3 and SlH4 as higher-level regulators that modulate Wo-dependent genetic networks governing tomato trichome diversity and structure.
“This discovery reveals how two zinc finger proteins cooperate with a master regulator to shape tomato trichomes,” said Dr. Jin-Ho Kang, corresponding author at Seoul National University. “By uncovering how SlH3 and SlH4 interact with Woolly to fine-tune downstream gene expression, we’ve bridged a major knowledge gap in multicellular trichome biology. This work highlights how complex regulatory partnerships govern the formation of specialized plant surface structures, which are essential for both stress tolerance and metabolic diversity.”
The elucidation of the SlH3–SlH4–Wo regulatory module opens new possibilities for engineering trichome traits in tomato and other crops. By manipulating these genes, breeders could enhance the production of glandular trichomes that secrete defense-related metabolites or modify trichome density for improved pest resistance. Furthermore, this discovery provides a valuable framework for exploring zinc finger–mediated transcriptional networks across plant species. Future research may leverage this pathway to design crops with optimized natural defense mechanisms, reducing dependence on chemical pesticides while maintaining productivity under challenging environmental conditions.
###
References
DOI
Original Source URL
https://doi.org/10.1093/hr/uhaf008
Funding information
This work was supported by grants from the New Breeding Technologies Development Program (RS-2024-00322125 and RS-2024-00322053) and Basic Science Research Program (NRF-2022R1A2C1008643) of the Republic of Korea.
About Horticulture Research
Horticulture Research is an open access journal of Nanjing Agricultural University and ranked number one in the Horticulture category of the Journal Citation Reports ™ from Clarivate, 2024. The journal is committed to publishing original research articles, reviews, perspectives, comments, correspondence articles and letters to the editor related to all major horticultural plants and disciplines, including biotechnology, breeding, cellular and molecular biology, evolution, genetics, inter-species interactions, physiology, and the origination and domestication of crops.
Disclaimer: AAAS and EurekAlert! are not responsible for the accuracy of news releases posted to EurekAlert! by contributing institutions or for the use of any information through the EurekAlert system.