Jenn Hoskins
11th December, 2025

High-efficiency genetic transformation of protoplasts from Safflower (Carthamus tinctorius) flowers was achieved using tissue from early flowering stages (2 days), establishing an optimal window for successful gene expression studies.
Key Findings
- Researchers identified CtMYB1, a key gene in safflower plants, crucial for flavonoid production, specifically in the flower petals
- Increasing CtMYB1 activity boosted expression of genes CtC4H2, CtF3H4, and CtHCT12 involved in flavonoid creation
- Suppressing CtMYB1 reduced the amount of Hydroxysafflor yellow A (HSYA), a major flavonoid, and lightened petal color
The study began with an investigation into the genetic makeup of safflower, focusing on genes similar to those known to control flavonoid synthesis in other plants. The team identified CtMYB1, a gene belonging to the MYB family of proteins. MYB proteins are transcription factors – essentially, they are proteins that control which genes are turned on or off[2]. In plants, these proteins are often central to regulating development, metabolism, and responses to environmental stresses.
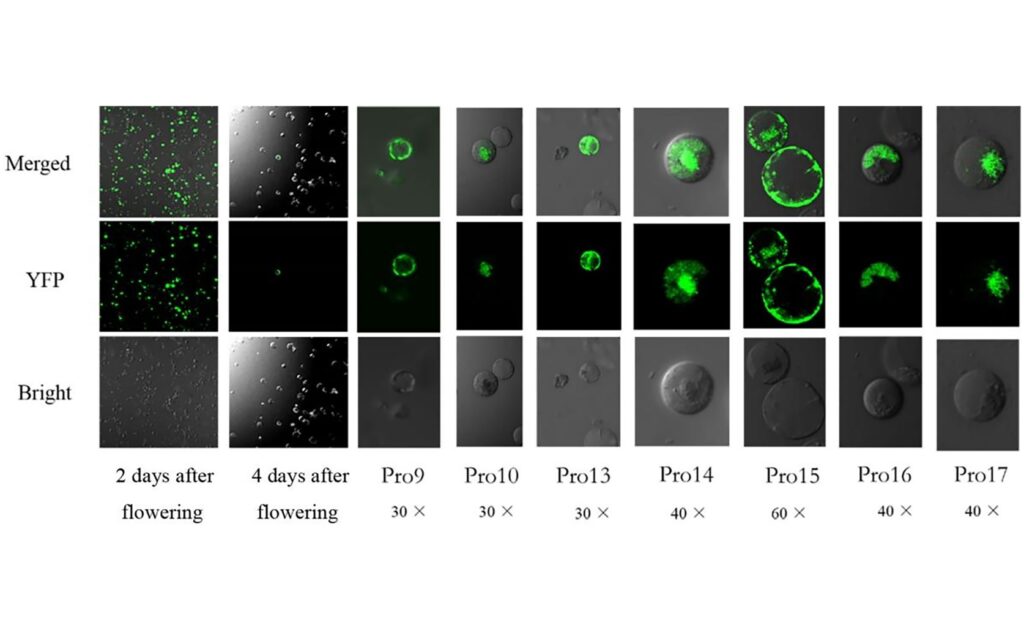
To understand CtMYB1’s function, the researchers needed a way to study gene activity directly within safflower cells. They successfully developed a method to introduce genes into safflower flower protoplasts – plant cells that have had their cell walls removed – allowing them to observe the effects of CtMYB1 on other genes. Simultaneously, they established a virus-induced gene silence (VIGS) technique. VIGS involves using a modified virus to temporarily ‘silence’ or deactivate specific genes within the plant, allowing researchers to observe the consequences of gene loss.
The protoplast experiments revealed that introducing CtMYB1 into safflower flower cells led to increased activity in several genes involved in flavonoid biosynthesis, specifically CtC4H2, CtF3H4, and CtHCT12. This indicated that CtMYB1 acts as an activator, promoting the production of these key enzymes. Conversely, using VIGS to suppress CtMYB1 resulted in paler petals and a reduction in the amount of Hydroxysafflor yellow A (HSYA), a major flavonoid component in safflower. These findings strongly suggest that CtMYB1 is essential for normal flavonoid production.
To confirm that CtMYB1 directly controls these biosynthesis genes, the researchers performed yeast one-hybrid experiments. This technique demonstrates whether a protein binds to specific DNA sequences. The results showed that CtMYB1 could bind to the promoters of CtC4H2, CtF3H4, and CtHCT12 – the regions of DNA that control gene activity. Further analysis using a Biacore system pinpointed a specific DNA sequence, CAACCA, on the promoters to which CtMYB1 binds. This precise binding site explains how CtMYB1 activates flavonoid biosynthesis genes.
These findings build upon previous research demonstrating the importance of polyphenols and serotonin derivatives found in safflower seed extract (SSE) for improving arterial health[3]. While that study focused on the effects of consuming SSE, the current research sheds light on the underlying mechanisms within the plant itself. Specifically, it identifies a key regulator of flavonoid production, the compounds responsible for many of those observed health benefits. The development of the flower protoplast expression system and VIGS technique in safflower provides powerful tools for further investigation into the genes involved in the production of active compounds within the plant, and for potentially enhancing flavonoid content in future crops.
References
Main Study
1) CtMYB1 regulates flavonoid biosynthesis in safflower flower by binding the CAACCA elements
Published 10th December, 2025
https://doi.org/10.1371/journal.pone.0337921
Related Studies
Related Articles